About group analyses
Group analysis in proteomics refers to the systematic comparison of protein abundance across different biological samples or conditions. This process involves grouping samples based on specific criteria—such as treatment conditions, disease states, or genetic backgrounds—and analyzing the differences in protein abundance within these groups.
Group analysis enables researchers to uncover the molecular basis of biological processes and disease states, potentially leading to the identification of biomarkers and therapeutic targets.
To run a group analysis, the original analysis results must have at least one category with two defined groups.
You can add or modify conditions in your Sample Description File or by editing samples in relevant plates in the Plates tab.
When you set up a group analysis, you must specify:
-
the data type: protein or peptide
-
the statistical test: T test or Wilcoxon Rank-Sum Test
-
a category and the two groups within it to compare (more)
You can also specify optional settings including:
-
filters to apply to sub-groups (more)
-
individual samples to exclude
-
quality thresholds where results that have undetected values by set number or percentage values can be excluded from the analysis (more)
-
a normalization method to reduce data variability that may arise from differences in sample preparation, instrumentation, or other technical factors: none, MaxLFQ, or median normalization
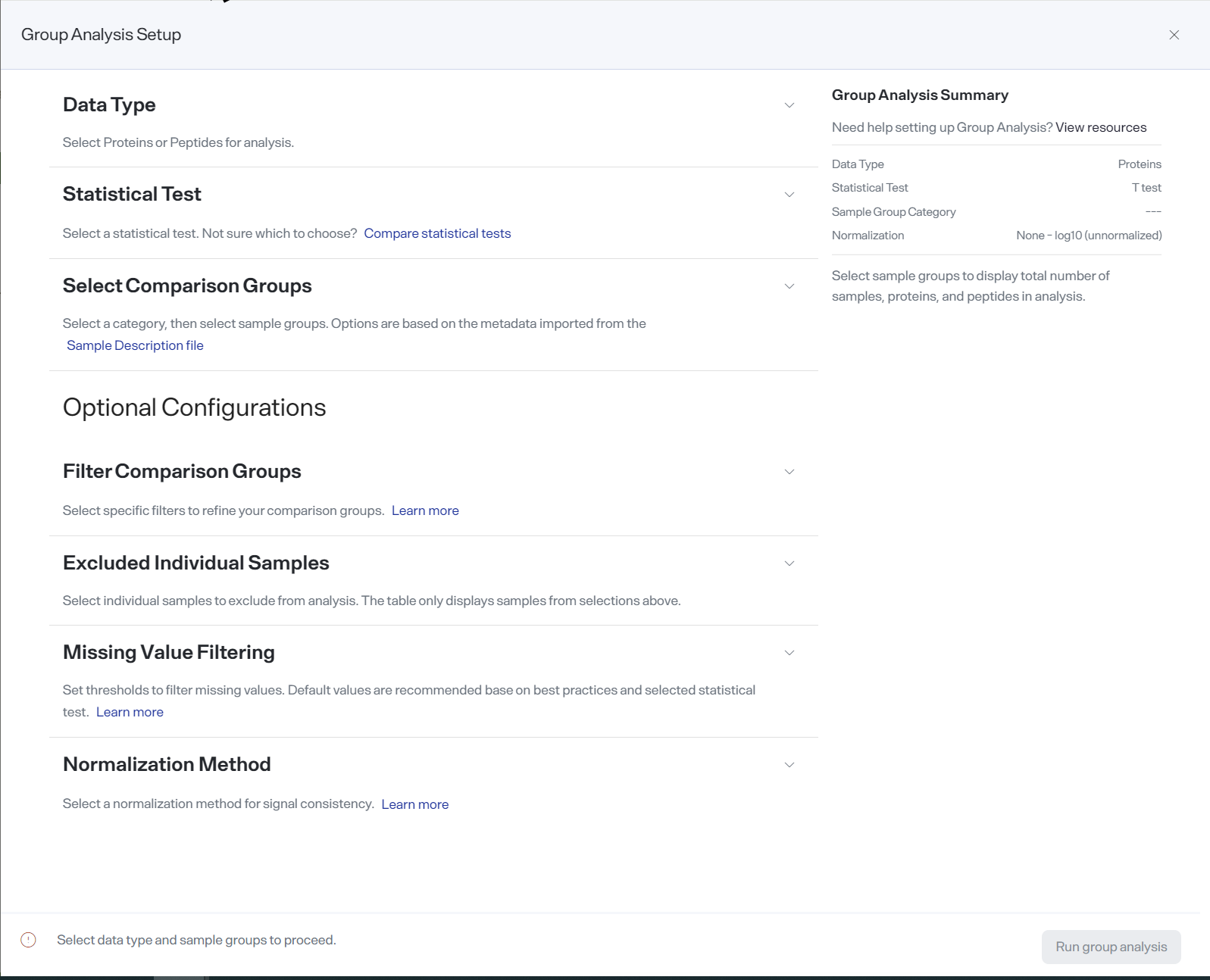
The Group Analysis Setup screen.
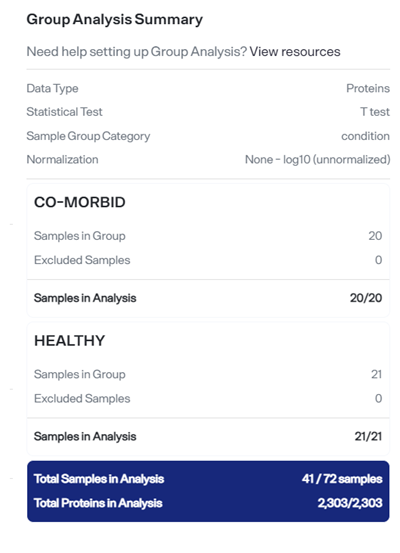
As you make selections, the Group Analysis Setup page displays real-time values, such as number of samples to be evaluated, based on the settings you've chosen.
The Group Analysis Summary section on the setup screen